What is the significance of genetic variants to those that carry them?

How do we determine the significance of genetic variants?
We are driven to understand how genetics shapes an individual's risk for disease or adverse outcomes. The Kroncke Lab pairs high-throughput assays, CRISPR-edited model systems, and statistical modeling to map how ion channel variants influence disease penetrance. Our team integrates functional data with genomic interpretation to clarify variants of uncertain significance in genes such as KCNH2 and SCN5A and to identify how polygenic background modifies expressivity in induced pluripotent stem cell-derived cardiomyocytes.
See this talk on YouTube for an overview of our research program.
What is our progress to date?
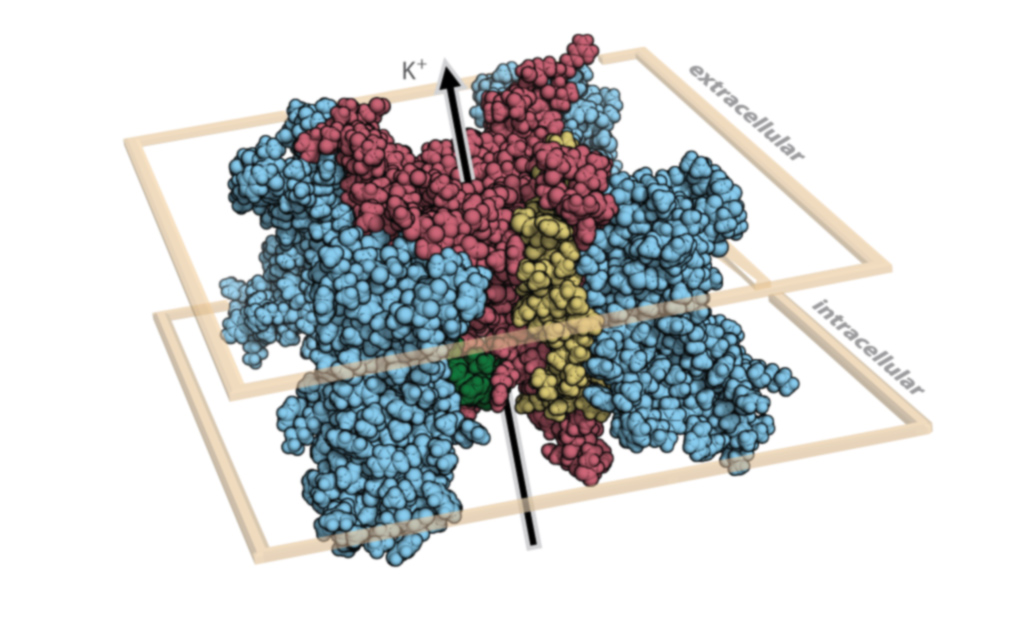
Above is a video highlighting an intersection of protein structure and clinical presentation, in this case for the sodium ion channel Nav1.5 (gene SCN5A). Variants associated with Brugada syndrome (BrS1, blue), type 3 long QT syndrome (LQT3, red), or unaffected carriers (gold) are shown as spheres. This representation suggests the utility of using structure and residue annotation to understand mechanism and generate predictions about yet uncharacterized variants found in structured regions.
Our patented Bayesian Method to Estimate Variant-Induced Disease Penetrance (US-20220406461-A1) and the VariantBrowser.org portal translate these insights into tools that clinicians and researchers worldwide rely on daily.
Integrating genetics, quantitative models, and high-throughput biology
What can help you determine the significance of genetic variants?
A list of useful resources for those trying to determine the significance of genetic variants.
Who does this work?
Current and past group members